PUBLICATIONS
JansenLab papersLars´ postdoc papers
Lars´ pre-PhD papers
Lars´ even earlier Papers
JansenLab papers
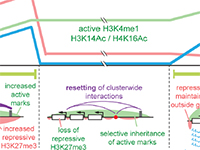
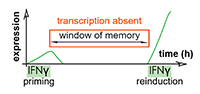
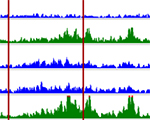 Pawel Mikulski, Sahar S.H. Tehrani, Anna Kogan, Izma Abdul-Zani, Emer Shell, Brent J. Ryan and Lars E.T. Jansen (2023) Transcriptional memory is conferred by combined heritable maintenance and local removal of selective chromatin modifications BioRxiv Preprint
Pawel Mikulski, Sahar S.H. Tehrani, Anna Kogan, Izma Abdul-Zani, Emer Shell, Brent J. Ryan and Lars E.T. Jansen (2023) Transcriptional memory is conferred by combined heritable maintenance and local removal of selective chromatin modifications BioRxiv Preprint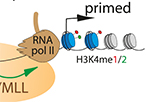 Sahar SH Tehrani, Anna Kogan, Pawel Mikulski and Lars ET Jansen (2023) Remembering foods and foes: emerging principles of transcriptional memory Nature, Cell Death & Differentiation
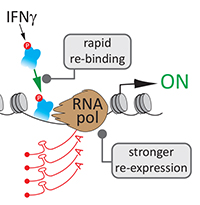
Sahar SH Tehrani, Anna Kogan, Pawel Mikulski and Lars ET Jansen (2023) Remembering foods and foes: emerging principles of transcriptional memory Nature, Cell Death & Differentiation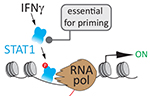 Sahar SH Tehrani, Pawel Mikulski, Izma Abdul-Zani, João F Mata, Wojciech Siwek and Lars ET Jansen (2023) STAT1 is required to establish but not maintain IFNγ-induced transcriptional memory EMBO Journal, e112259
Sahar SH Tehrani, Pawel Mikulski, Izma Abdul-Zani, João F Mata, Wojciech Siwek and Lars ET Jansen (2023) STAT1 is required to establish but not maintain IFNγ-induced transcriptional memory EMBO Journal, e112259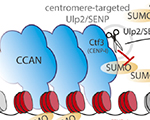 Sebastiaan J.W. van den Berg and Lars E.T. Jansen (2023) SUMO control of centromere
homeostasis Front. Cell Dev. Biol., doi.org/10.3389/fcell.2023.1193192
Sebastiaan J.W. van den Berg and Lars E.T. Jansen (2023) SUMO control of centromere
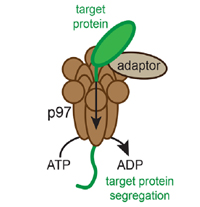
homeostasis Front. Cell Dev. Biol., doi.org/10.3389/fcell.2023.1193192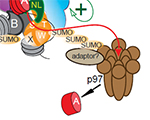 Sebastiaan J.W. van den Berg, Samuel East, Sreyoshi Mitra and Lars E.T. Jansen (2023) p97/VCP drives turnover of SUMOylated centromeric CCAN proteins and CENP-A MBoC, doi.org/10.1091/mbc.E23-01-0035
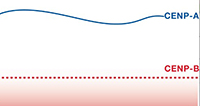
Sebastiaan J.W. van den Berg, Samuel East, Sreyoshi Mitra and Lars E.T. Jansen (2023) p97/VCP drives turnover of SUMOylated centromeric CCAN proteins and CENP-A MBoC, doi.org/10.1091/mbc.E23-01-0035 Dragan Stajic and Lars E. T. Jansen (2021) Empirical evidence for epigenetic inheritance driving evolutionary adaptation Phil. Trans. R. Soc., B3762020012120200121
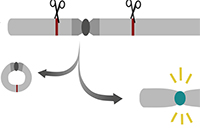
Dragan Stajic and Lars E. T. Jansen (2021) Empirical evidence for epigenetic inheritance driving evolutionary adaptation Phil. Trans. R. Soc., B3762020012120200121 Marina Murillo-Pineda, Luis P. Valente, Marie Dumont, João F. Mata, Daniele Fachinetti and Lars E.T. Jansen (2021) Induction of spontaneous human neocentromere formation and long-term maturation. Journal of Cell Biology, 220 (3): e202007210
Marina Murillo-Pineda, Luis P. Valente, Marie Dumont, João F. Mata, Daniele Fachinetti and Lars E.T. Jansen (2021) Induction of spontaneous human neocentromere formation and long-term maturation. Journal of Cell Biology, 220 (3): e202007210Highlighed by Ben Carty and Elaine Dunleavy
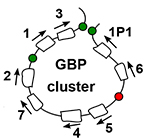 Wojciech Siwek, Sahar S.H. Tehrani, João F. Mata and Lars E.T. Jansen (2020) Activation of clustered IFNg target genes drives cohesin-controlled transcriptional memory. Molecular Cell, doi.org/10.1016/j.molcel.2020.10.005
Wojciech Siwek, Sahar S.H. Tehrani, João F. Mata and Lars E.T. Jansen (2020) Activation of clustered IFNg target genes drives cohesin-controlled transcriptional memory. Molecular Cell, doi.org/10.1016/j.molcel.2020.10.005 Inês Milagre, Carolina Pereira, Raquel Oliveira and Lars E.T. Jansen (2020) Reprogramming of Human Cells to Pluripotency Induces CENP-A Chromatin Depletion. Open Biology,10: 200227
Inês Milagre, Carolina Pereira, Raquel Oliveira and Lars E.T. Jansen (2020) Reprogramming of Human Cells to Pluripotency Induces CENP-A Chromatin Depletion. Open Biology,10: 200227 Sebastiaan J. W. van den Berg and Lars E.T. Jansen (2020) Centromeres: Genetic input to calibrate an epigenetic feedback loop EMBO Journal, e106638
Sebastiaan J. W. van den Berg and Lars E.T. Jansen (2020) Centromeres: Genetic input to calibrate an epigenetic feedback loop EMBO Journal, e106638 Sreyoshi Mitra, Bharath Srinivasan and Lars E.T. Jansen (2020) Stable inheritance of CENP-A chromatin: Inner strength versus dynamic control Journal of Cell Biology, 219 (10): e202005099
Sreyoshi Mitra, Bharath Srinivasan and Lars E.T. Jansen (2020) Stable inheritance of CENP-A chromatin: Inner strength versus dynamic control Journal of Cell Biology, 219 (10): e202005099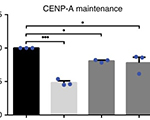 Georg O. M. Bobkov, Anming Huang, Sebastiaan J. W. van den Berg, Sreyoshi Mitra, Eduard Anselm1, Vasiliki Lazou1, Sarah Schunter, Regina Feederle, Axel Imhof, Alexandra Lusser, Lars E. T. Jansen and Patrick Heun (2020) Spt6 is a maintenance factor for centromeric
CENP-A Nature Communications, 11: 2020
Georg O. M. Bobkov, Anming Huang, Sebastiaan J. W. van den Berg, Sreyoshi Mitra, Eduard Anselm1, Vasiliki Lazou1, Sarah Schunter, Regina Feederle, Axel Imhof, Alexandra Lusser, Lars E. T. Jansen and Patrick Heun (2020) Spt6 is a maintenance factor for centromeric
CENP-A Nature Communications, 11: 2020
 Marina Murillo-Pineda and Lars E.T. Jansen (2020) Genetics, epigenetics and back again: Lessons learned from neocentromeres.
Experimental Cell Research doi.org/10.1016/j.yexcr.2020.111909
Marina Murillo-Pineda and Lars E.T. Jansen (2020) Genetics, epigenetics and back again: Lessons learned from neocentromeres.
Experimental Cell Research doi.org/10.1016/j.yexcr.2020.111909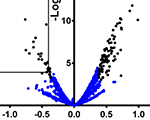 Sreyoshi Mitra, Dani L. Bodor, Ana F. David, Izma Abdul-Zani João F. Mata, Beate Neumann, Sabine Reither, Christian Tischer and Lars E.T. Jansen (2020) Genetic screening identifies a SUMO protease dynamically maintaining centromeric chromatin.
Nature Communications, 11: 501
Sreyoshi Mitra, Dani L. Bodor, Ana F. David, Izma Abdul-Zani João F. Mata, Beate Neumann, Sabine Reither, Christian Tischer and Lars E.T. Jansen (2020) Genetic screening identifies a SUMO protease dynamically maintaining centromeric chromatin.
Nature Communications, 11: 501
 Dragan Stajic, Lília Perfeito and Lars E.T. Jansen (2019) Epigenetic gene silencing alters the mechanisms and rate of evolutionary adaptation. Nature Ecology & Evolution, 3: 491-8
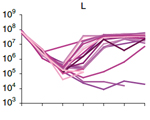
Dragan Stajic, Lília Perfeito and Lars E.T. Jansen (2019) Epigenetic gene silencing alters the mechanisms and rate of evolutionary adaptation. Nature Ecology & Evolution, 3: 491-8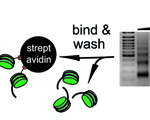 Wojciech Siwek, Mariluz Gómez-Rodríguez, Daniel Sobral, Ivan R. Corrêa Jr and Lars E.T. Jansen (2018) time-ChIP: A method to determine long-term locus-specific nucleosome inheritance. Methods Mol. Biol., 1832: 131-58
Wojciech Siwek, Mariluz Gómez-Rodríguez, Daniel Sobral, Ivan R. Corrêa Jr and Lars E.T. Jansen (2018) time-ChIP: A method to determine long-term locus-specific nucleosome inheritance. Methods Mol. Biol., 1832: 131-58
 Ana Stankovic and Lars E.T Jansen (2017) Quantitative Microscopy Reveals Centromeric Chromatin Stability, Size, and Cell Cycle Mechanisms to Maintain Centromere Homeostasis. Prog Mol Subcell Biol., 56: 139-162
Ana Stankovic and Lars E.T Jansen (2017) Quantitative Microscopy Reveals Centromeric Chromatin Stability, Size, and Cell Cycle Mechanisms to Maintain Centromere Homeostasis. Prog Mol Subcell Biol., 56: 139-162
 Ana Stankovic, Lucie Y. Guo, João F. Mata, Dani L. Bodor, Xing-Jun Cao, Aaron O. Bailey, Jeffrey Shabanowitz, Donald F. Hunt, Benjamin A. Garcia, Ben E. Black and Lars E.T Jansen (2017)
A dual inhibitory mechanism sufficient to maintain cell cycle restricted CENP-A assembly. Molecular Cell, 65: 231–246
Ana Stankovic, Lucie Y. Guo, João F. Mata, Dani L. Bodor, Xing-Jun Cao, Aaron O. Bailey, Jeffrey Shabanowitz, Donald F. Hunt, Benjamin A. Garcia, Ben E. Black and Lars E.T Jansen (2017)
A dual inhibitory mechanism sufficient to maintain cell cycle restricted CENP-A assembly. Molecular Cell, 65: 231–246 Aimee M Deaton, Mariluz Gomez-Rodriguez, Jakub Mieczkowski, Michael Y Tolstorukov, Sharmistha Kundu, Ruslan I Sadreyev, Lars E.T. Jansen and Robert E Kingston (2016)
Enhancer regions show high histone H3.3 turnover that changes during differentiation eLife, 5:e15316
Aimee M Deaton, Mariluz Gomez-Rodriguez, Jakub Mieczkowski, Michael Y Tolstorukov, Sharmistha Kundu, Ruslan I Sadreyev, Lars E.T. Jansen and Robert E Kingston (2016)
Enhancer regions show high histone H3.3 turnover that changes during differentiation eLife, 5:e15316
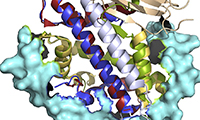
 Samantha J Falk, Lucie Y Guo, Nikolina Sekulic, Evan M Smoak, Tomoyasu Mani, Glennis A Logsdon, Kushol Gupta, Lars E T Jansen, Gregory D Van Duyne, Sergei A Vinogradov, Michael A Lampson and Ben E Black (2015)
CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science, 348: 699-703
Samantha J Falk, Lucie Y Guo, Nikolina Sekulic, Evan M Smoak, Tomoyasu Mani, Glennis A Logsdon, Kushol Gupta, Lars E T Jansen, Gregory D Van Duyne, Sergei A Vinogradov, Michael A Lampson and Ben E Black (2015)
CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science, 348: 699-703
 Dani L. Bodor, João F. Mata, Mikhail Sergeev, Ana Filipa David, Kevan J. Salimian, Tanya Panchenko, Don W. Cleveland, Ben E. Black, Jagesh V. Shah and Lars E.T. Jansen (2014)
The quantitative architecture of centromeric chromatin. eLife, 2014;3:e02137
Dani L. Bodor, João F. Mata, Mikhail Sergeev, Ana Filipa David, Kevan J. Salimian, Tanya Panchenko, Don W. Cleveland, Ben E. Black, Jagesh V. Shah and Lars E.T. Jansen (2014)
The quantitative architecture of centromeric chromatin. eLife, 2014;3:e02137
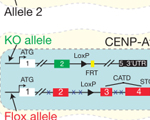 Daniele Fachinetti, H. Diego Folco, Yael Nechemia-Arbely, Luis P. Valente, Kristen Nguyen, Alex J. Wong, Quan Zhu, Andrew J. Holland, Arshad Desai, Lars E. T. Jansen and Don W. Cleveland (2013)
A two-step mechanism for epigenetic specification of centromere identity and function. Nature Cell Biol., 15: 1056-66.
Daniele Fachinetti, H. Diego Folco, Yael Nechemia-Arbely, Luis P. Valente, Kristen Nguyen, Alex J. Wong, Quan Zhu, Andrew J. Holland, Arshad Desai, Lars E. T. Jansen and Don W. Cleveland (2013)
A two-step mechanism for epigenetic specification of centromere identity and function. Nature Cell Biol., 15: 1056-66.
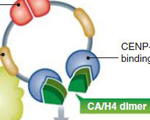 Dani L. Bodor and Lars E.T. Jansen (2013) How two become one: HJURP dimerization drives CENP-A assembly. EMBO J., 32: 2090-2.
Dani L. Bodor and Lars E.T. Jansen (2013) How two become one: HJURP dimerization drives CENP-A assembly. EMBO J., 32: 2090-2.
William C. Earnshaw et al., (2013) Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome Res., 21: 101-6.
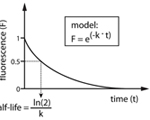 Dani L. Bodor, Luis P. Valente, João F. Mata, Ben E. Black and Lars E.T. Jansen (2013) Assembly in G1 phase and Long-Term Stability are Unique Intrinsic Features of CENP-A nucleosomes.
Mol. Biol. Cell., 24: 923-932
Dani L. Bodor, Luis P. Valente, João F. Mata, Ben E. Black and Lars E.T. Jansen (2013) Assembly in G1 phase and Long-Term Stability are Unique Intrinsic Features of CENP-A nucleosomes.
Mol. Biol. Cell., 24: 923-932
 Ana Stankovic and Lars E.T. Jansen (2013) Reductionism at the vertebrate kinetochore. Journal of Cell Biology, 200: 7-8
Ana Stankovic and Lars E.T. Jansen (2013) Reductionism at the vertebrate kinetochore. Journal of Cell Biology, 200: 7-8
Mariluz Gómez-Rodríguez and Lars E.T. Jansen (2013) Basic properties of epigenetic systems: lessons from the centromere. Curr Opin Genet Dev., 23: 219-27
 Luis P. Valente, Mariana C.C. Silva and Lars E.T. Jansen (2012)Temporal control of epigenetic centromere specification. Chromosome Research, 20:481-92
Luis P. Valente, Mariana C.C. Silva and Lars E.T. Jansen (2012)Temporal control of epigenetic centromere specification. Chromosome Research, 20:481-92
 Dani L. Bodor, Mariluz Gómez Rodríguez, Nuno Moreno and Lars E.T. Jansen (2012) Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Current Protocols in Cell Biology, Unit 8.8.1-8.8.34, June 2012
Dani L. Bodor, Mariluz Gómez Rodríguez, Nuno Moreno and Lars E.T. Jansen (2012) Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Current Protocols in Cell Biology, Unit 8.8.1-8.8.34, June 2012
 João F. Mata, Telma Lopes, Rui Gardner and Lars E.T. Jansen (2012) A rapid FACS-based strategy to isolate human gene knockin and knockout clones. PLoS ONE, 7:e32646
João F. Mata, Telma Lopes, Rui Gardner and Lars E.T. Jansen (2012) A rapid FACS-based strategy to isolate human gene knockin and knockout clones. PLoS ONE, 7:e32646
 Lars E.T. Jansen (2012) Sowing the seeds of centromeres. Science, 335: 299-300
Lars E.T. Jansen (2012) Sowing the seeds of centromeres. Science, 335: 299-300
 Mariana C.C. Silva, Dani L. Bodor, Madison E. Stellfox, Nuno M.C. Martins, Helfrid Hochegger, Daniel R. Foltz and Lars E.T. Jansen (2012) Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Developmental Cell, 22: 52-63
Mariana C.C. Silva, Dani L. Bodor, Madison E. Stellfox, Nuno M.C. Martins, Helfrid Hochegger, Daniel R. Foltz and Lars E.T. Jansen (2012) Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Developmental Cell, 22: 52-63 (Highlighed by Kaitlin M. Stimpson and Beth A. Sullivan)
 Dominique Ray Gallet, Adam Woolfe, Isabelle Vassias, Céline Pellentz, Nicolas Lacoste, Aastha Puri, David C. Schultz, Nikolay A. Pchelintsev, Peter D. Adams, Lars E.T. Jansen and Geneviève Almouzni (2011)
Dynamics of histone H3 deposition in vivo reveal a gap filling mechanism for H3.3 to maintain chromatin integrity. Molecular Cell, 44: 928-41
Dominique Ray Gallet, Adam Woolfe, Isabelle Vassias, Céline Pellentz, Nicolas Lacoste, Aastha Puri, David C. Schultz, Nikolay A. Pchelintsev, Peter D. Adams, Lars E.T. Jansen and Geneviève Almouzni (2011)
Dynamics of histone H3 deposition in vivo reveal a gap filling mechanism for H3.3 to maintain chromatin integrity. Molecular Cell, 44: 928-41
 Jan H. Bergmann, Mariluz Gómez Rodríguez, Nuno M. C. Martins, Hiroshi Kimura, David A. Kelly, Hiroshi Masumoto, Vladimir Larionov, Lars E. T. Jansen and William C Earnshaw (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO Journal, 30:328-40
Jan H. Bergmann, Mariluz Gómez Rodríguez, Nuno M. C. Martins, Hiroshi Kimura, David A. Kelly, Hiroshi Masumoto, Vladimir Larionov, Lars E. T. Jansen and William C Earnshaw (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO Journal, 30:328-40
 Ben E. Black, Lars E.T. Jansen, Daniel R. Foltz and Don W. Cleveland (2010) Centromere Identity, Function, and Epigenetic Propagation across Cell Divisions. Cold Spring Harb Symp Quant Biol, 75:403-18
Ben E. Black, Lars E.T. Jansen, Daniel R. Foltz and Don W. Cleveland (2010) Centromere Identity, Function, and Epigenetic Propagation across Cell Divisions. Cold Spring Harb Symp Quant Biol, 75:403-18
 Mariana C.C. Silva and Lars E.T. Jansen (2009) At the right place at the right time: Novel CENP-A binding proteins shed light on centromere assembly. Chromosoma, 118: 567-74
Mariana C.C. Silva and Lars E.T. Jansen (2009) At the right place at the right time: Novel CENP-A binding proteins shed light on centromere assembly. Chromosoma, 118: 567-74
 Christopher W. Carroll, Mariana C.C. Silva, Kristina M. Godek, Lars E.T. Jansen and Aaron F. Straight (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nature Cell Biol., 11, 896-902
Christopher W. Carroll, Mariana C.C. Silva, Kristina M. Godek, Lars E.T. Jansen and Aaron F. Straight (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nature Cell Biol., 11, 896-902
Lars´ Postdoc Papers
Ludwig Institute for Cancer Research, La Jolla, CA
 Alexander Samoshkin, Alexei Arnaoutov, Lars E. T. Jansen, Ilia Ouspenski, Louis Dye, Tatiana Karpova, James McNally, Mary Dasso, Don W. Cleveland and Alexander Strunnikov (2009). Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS One, 4:e6831.
Alexander Samoshkin, Alexei Arnaoutov, Lars E. T. Jansen, Ilia Ouspenski, Louis Dye, Tatiana Karpova, James McNally, Mary Dasso, Don W. Cleveland and Alexander Strunnikov (2009). Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS One, 4:e6831.
 Daniel R. Foltz, Lars E.T. Jansen, Aaron O. Bailey, John R. Yates III, Emily A. Bassett, Stacey Wood, Ben E. Black and Don W. Cleveland (2009) Centromere-Specific Assembly of CENP-A Nucleosomes Is Mediated by HJURP. Cell, 137, 472-84
Daniel R. Foltz, Lars E.T. Jansen, Aaron O. Bailey, John R. Yates III, Emily A. Bassett, Stacey Wood, Ben E. Black and Don W. Cleveland (2009) Centromere-Specific Assembly of CENP-A Nucleosomes Is Mediated by HJURP. Cell, 137, 472-84
 Lars E.T. Jansen, Ben E. Black, Daniel R. Foltz, and Don W. Cleveland (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol., 176, 795-805
Lars E.T. Jansen, Ben E. Black, Daniel R. Foltz, and Don W. Cleveland (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol., 176, 795-805
Highlighed by Christopher W. Carroll and Aaron F. Straight and Mitch Leslie.
 Ben E. Black*, Lars E.T. Jansen*, Paul S. Maddox, Daniel R. Foltz, Arshad B. Desai, Jagesh V. Shah and Don W. Cleveland (2007) Centromere Identity Maintained by Nucleosomes Assembled with Histone H3 Containing the CENP-A Targeting Domain. Molecular Cell, 25, 309-22
*equal contributors
Ben E. Black*, Lars E.T. Jansen*, Paul S. Maddox, Daniel R. Foltz, Arshad B. Desai, Jagesh V. Shah and Don W. Cleveland (2007) Centromere Identity Maintained by Nucleosomes Assembled with Histone H3 Containing the CENP-A Targeting Domain. Molecular Cell, 25, 309-22
*equal contributors
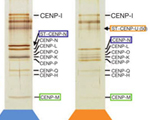 Daniel R. Foltz, Lars E.T. Jansen, Ben E. Black, Aaron O. Bailey, John R. Yates III and Don W. Cleveland (2006) The human CENP-A centromeric nucleosome-associated complex. Nature Cell Biology, 8, 458-69
Daniel R. Foltz, Lars E.T. Jansen, Ben E. Black, Aaron O. Bailey, John R. Yates III and Don W. Cleveland (2006) The human CENP-A centromeric nucleosome-associated complex. Nature Cell Biology, 8, 458-69
Lars´ pre-PhD Papers
Leiden Institute for Chemistry, Leiden University
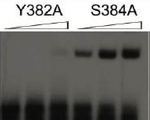 Masakazu Kobayashi, Francis Figaroa, Nico Meeuwenoord, Lars E.T. Jansen and Gregg Siegal (2006) Characterization of the DNA binding and structural properties of the BRCT region of human replication factor C p140 subunit.
J Biol Chem., 281, 4308-17
Masakazu Kobayashi, Francis Figaroa, Nico Meeuwenoord, Lars E.T. Jansen and Gregg Siegal (2006) Characterization of the DNA binding and structural properties of the BRCT region of human replication factor C p140 subunit.
J Biol Chem., 281, 4308-17
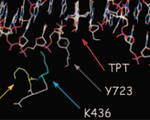 Robert C. A. M. van Waardenburg, Laurina A. de Jong, Maria A. J. van Eijndhoven, Caroline Verseyden, Dick Pluim, Lars E.T. Jansen, Mary-Ann Bjornsti, and Jan H. M. Schellens (2004) Platinated DNA Adducts Enhance Poisoning of DNA Topoisomerase I by Camptothecin. J. Biol. Chem., 279, 54502-9.
Robert C. A. M. van Waardenburg, Laurina A. de Jong, Maria A. J. van Eijndhoven, Caroline Verseyden, Dick Pluim, Lars E.T. Jansen, Mary-Ann Bjornsti, and Jan H. M. Schellens (2004) Platinated DNA Adducts Enhance Poisoning of DNA Topoisomerase I by Camptothecin. J. Biol. Chem., 279, 54502-9.
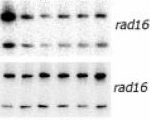 Lars E.T. Jansen, Ana I. Belo, Rinske Hulsker and Jaap Brouwer (2002) Transcription elongation factor Spt4 mediates loss of phosphorylated RNA polymerase II transcription in response to DNA damage. Nucleic Acids Res., 30, 3532-9.
Lars E.T. Jansen, Ana I. Belo, Rinske Hulsker and Jaap Brouwer (2002) Transcription elongation factor Spt4 mediates loss of phosphorylated RNA polymerase II transcription in response to DNA damage. Nucleic Acids Res., 30, 3532-9.
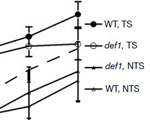 Elies C. Woudstra, Chris Gilbert, Jane Fellows, Lars E.T. Jansen, Jaap Brouwer, Hediye Erdjument-Bromage, Paul Tempst and Jesper Q. Svejstrup (2002) A Rad26-Def1 complex co-ordinates repair and RNA polymerase II proteolysis in response to DNA damage. Nature. 415, 929-33.
Elies C. Woudstra, Chris Gilbert, Jane Fellows, Lars E.T. Jansen, Jaap Brouwer, Hediye Erdjument-Bromage, Paul Tempst and Jesper Q. Svejstrup (2002) A Rad26-Def1 complex co-ordinates repair and RNA polymerase II proteolysis in response to DNA damage. Nature. 415, 929-33.
 Lars E.T. Jansen, Hans den Dulk, Rosalba M. Brouns, Martina de Ruijter, Jourica A. Brandsma and Jaap Brouwer (2000) Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J., 19, 6498-507.
Lars E.T. Jansen, Hans den Dulk, Rosalba M. Brouns, Martina de Ruijter, Jourica A. Brandsma and Jaap Brouwer (2000) Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J., 19, 6498-507.
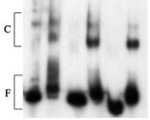 Lars E.T. Jansen, Richard A. Verhage and Jaap Brouwer (1998) Preferential binding of yeast Rad4-Rad23 complex to damaged DNA. J Biol Chem., 273, 33111-4.
Lars E.T. Jansen, Richard A. Verhage and Jaap Brouwer (1998) Preferential binding of yeast Rad4-Rad23 complex to damaged DNA. J Biol Chem., 273, 33111-4.
Lars´ even earlier Papers
Institute for Molecular Plant sciences, Leiden University
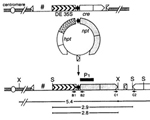 Annette C. Vergunst, Lars E.T. Jansen, Paul F. Fransz, J. Hans de Jong and Paul J.J. Hooykaas (2000) Cre/lox-mediated recombination in Arabidopsis: evidence for transmission of a translocation and a deletion event. Chromosoma. 109, 287-97.
Annette C. Vergunst, Lars E.T. Jansen, Paul F. Fransz, J. Hans de Jong and Paul J.J. Hooykaas (2000) Cre/lox-mediated recombination in Arabidopsis: evidence for transmission of a translocation and a deletion event. Chromosoma. 109, 287-97.
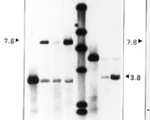 Annette C. Vergunst, Lars E.T. Jansen and Paul J. Hooykaas (1998) Site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana mediated by Cre recombinase. Nucleic Acids Res., 26, 2729-34.
Annette C. Vergunst, Lars E.T. Jansen and Paul J. Hooykaas (1998) Site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana mediated by Cre recombinase. Nucleic Acids Res., 26, 2729-34.




 In a dramatic departure from human centromeres and mammalian transcription we explored this key question in evolution in a wonderful collaboration with
In a dramatic departure from human centromeres and mammalian transcription we explored this key question in evolution in a wonderful collaboration with