RESEARCH
What is the basis for chromatin-based inheritance?
- Our Central Hypothesis
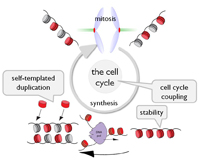
In addition to the primary DNA sequence, other cellular components can be heritbale if 1) they can survive through mitotic divisions 2) are self-templating in nature and 3) their duplication and transmission is coupled to cell division.
We work on aspects of all three of these themes
The Problem
DNA sequences that constitute the genome are replicated in a self-templating manner and transmitted through cell division resulting in fateful inheritance of genetic information.
In addition to the bare-bones genome, chromosome structure and patterns of gene expression are also maintained through mitotic and sometimes even meiotic divisions.
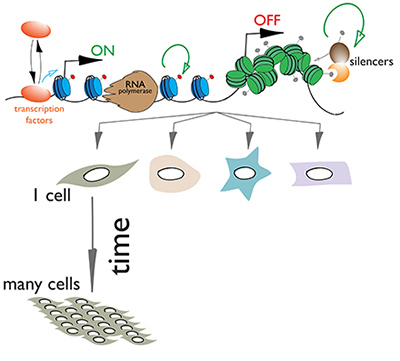
Although the basic mechanism of duplication and mitotic transmission of DNA sequences has been worked out decades ago, how other components, such as proteins, that are part of chromatin and govern gene activities and chromosome structure are maintained through cell division is not understood. We are interested in resolving this.
Our lab aims at three major questions
1. The Human Centromere
Centromeres are chromosomal loci that form the attachment site for spindle microtubules, driving chromosome segregation in mitosis. Centromeres are defined by a unique chromatin structure featuring the histone H3 variant CENP-A. Nucleosomes containing CENP-A are at the center of an epigenetic feedback loop where chromatin-bound CENP-A is sufficient to initiate self-templated inheritance of centromeres. We work on determining how CENP-A chromatin can be transmitted through cell division, how its assembly is restricted to the centromere and how inheritance is cell cycle controlled.
2. Transcriptional Memory
Cytokine signalling leads to gene induction. Strikingly, exposure to cytokines such as interferons can lead to long-term potentiation of transcription. Re-exposure of cells to the inducing signal leads to an enhanced response in primed cells, even after multiple cell division cycles.
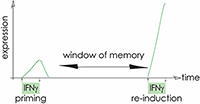
We aim to understand the mechanistic basis of transcriptional memory and define the role of chromatin, transcription factor networks and cytokine signalling.
We focus on chromatin, the protein DNA complex that packages DNA. Histone proteins, which make up the nucleosome may contribute to "memory" of prior gene expression.
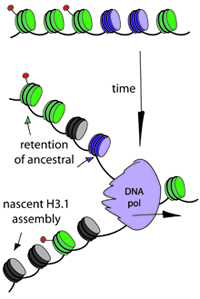 how is chromatin inherited across the cell cycle?
how is chromatin inherited across the cell cycle?
3. Yeast Population Epigenetics
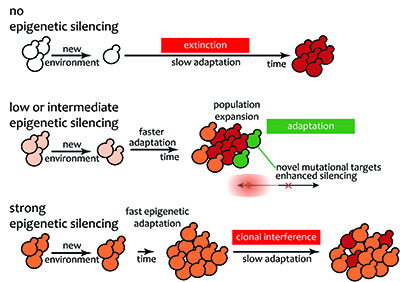
Gene silencing is maintained through cell division. In principle, any heritable trait that has a phenotypic consequence can be subject to selection. We explore the possibility that chromatin-based heritable gene silencing can change the rate and/or the mechanisms of adaptation in a population when gene silencing is under selection. We aim to understand how short-term epigenetic inheritance impacts on long-term adaptation in evolution.



 how is chromatin inherited across the cell cycle?
how is chromatin inherited across the cell cycle?
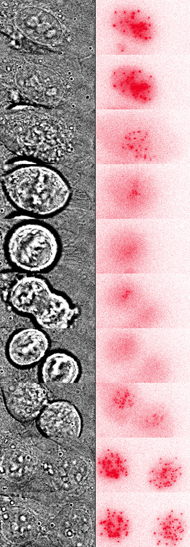
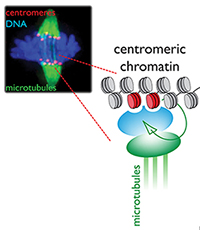
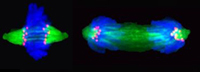 Much of our work focuses on the centromere, a unique chromatin domain connecting chromosomes to the mitotic spindle during mitosis. The centromere is maintained epigenetically through the propagation of a specialized chromatin structure that is not determined directly by the underlying DNA sequences. Instead, a unique histone H3 variant, CENP-A (red foci) marks the site of the centromere and is critical for its epigenetic propagation.
Much of our work focuses on the centromere, a unique chromatin domain connecting chromosomes to the mitotic spindle during mitosis. The centromere is maintained epigenetically through the propagation of a specialized chromatin structure that is not determined directly by the underlying DNA sequences. Instead, a unique histone H3 variant, CENP-A (red foci) marks the site of the centromere and is critical for its epigenetic propagation.
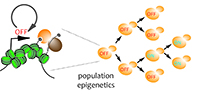 Yeast is a single celled organisms ideally suited for experimental evolution to discover how silent chromatin changes the way populations adapt to a novel environment.
Yeast is a single celled organisms ideally suited for experimental evolution to discover how silent chromatin changes the way populations adapt to a novel environment.